reference: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5411771/pdf/768.pdf
Summary
ABySS 1.0, was the first scalable de novo assembly tool that could assemble a human genome using short reads from high-throughput sequencing platform (Simpson et at 2009). Yet large amount of memory distributed across compute nodes (MPI) is required.
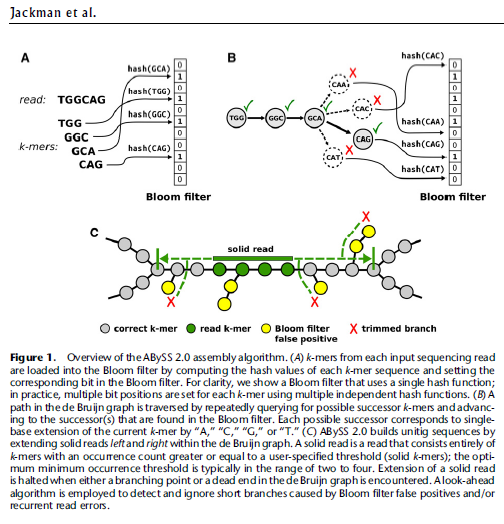
ABySS 2.0 implements algorithms that employ a Bloom filter, a probabilistic data structure, to represent a de Bruijn graph and reduce memory requirements by an order of magnitude
In this paper, the authors benchmarked ABySS 2.0 on a human genome assembly using a Genome in a Bottle data set of 250-bp Illumina paired-end. The final assembly yielded a NG50 (NGA50) scaffold contiguity of 3.5 (3.0) Mbp using <35 GB of RAM.