Overview
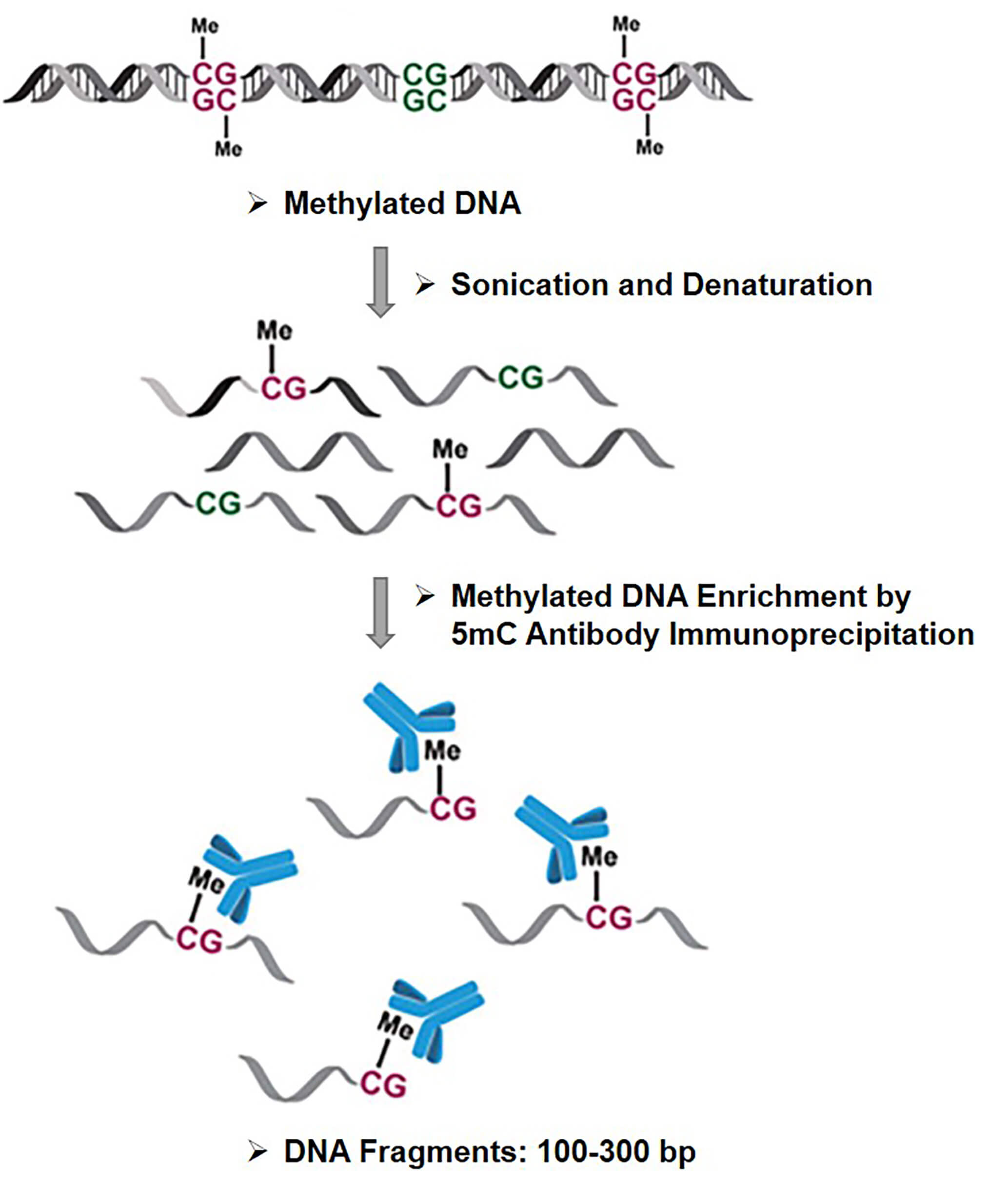
MeDIP (Methylated DNA immuniprecipitation) is an enrichment technique that uses antibodies specific to 5mC (methylated cytocines) to isolate methylated DNA fragments.
In brief, the DNA is denaturated to separate the double-strnaded DNA into single stranded DNA. This process enables the methyl-group to be accessible. Then, monoclonal or polyclonal antibodies with a binding epitope specific to methylated cytosines are incubated with the single-stranded DNA to promote interaction.
Applications/Limitations/advantages
The anti-5mC antibodies recognize and bind to methylated DNA irrespective of the regions making MeDIP interesting for whole genome screening. The MeDIP does not rely on bisulfite conversion or enzymatic digestion or size-selection, making this technique fast and relativelty cost effective as compared to other techniques. This is a useful technique when you are interested in determining methylated regions rather than methylation status at individual cytosines.
whole-genome bisulfite sequencing (WGBS) or reduced representation bisulfite sequencing (RRBS) require bisulfite conversion of unmethylated cytosines before the sequencing step. One drawback is that the majority of DNA is degraded. Input material need to be large as compared to MeDIP. (100ng compared to WGBS and RBBS, which require 2000 and 1000ng respectively).
One limitation is that while MeDIP non-specific binding of the anti-5mC is kept to a minimum, the anti-5mC antibody is biased towards highly methylated regions such as CpG islands. Low density regions may be missed.